Study investigates COVID-19 reinfection mechanism
25/05/2021
Solange Argenta (Fiocruz Pernambuco) and Bruno Moraes (Genomics Network)
The rapid spread of some Sars-CoV-2 variants has been associated with reports of COVID-19 reinfection, based, for example, on the P.1 high prevalence in the first two months after its emergence. Given this scenario, researchers from Fiocruz Pernambuco and Foundation's Genomics Network members sought to verify what the real mechanisms behind reinfection might be. The initial hypothesis was that a greater competence of the new variants would bind to human host cells, but the result pointed in a different direction. According to the study coordinator and Fiocruz Pernambuco researcher, Roberto Lins, it was observed that the antibodies generated in the pandemic first wave could not efficiently neutralize the variants of concern studied. These new mutations the virus has undergone have in fact provided it with the ability to evade the body's immune response.
The results are in the paper Immune Evasion of Sars-CoV-2 Variants of Concern is Driven by Low Affinity to Neutralizing Antibodies, which has just been published in the scientific journal Chemical Communications. The authors, besides Roberto Lins himself, are the Fiocruz Pernambuco researcher Gabriel Wallau; the chemistry doctoral students Emerson Gonçalves Moreira and Matheus Ferraz; and the post-doctoral fellow in chemistry Danilo Coelho (all with academic degrees from the Federal University of Pernambuco, UFPE, and working at Fiocruz Pernambuco in research related to monitoring the Genomics Network).
Methodology
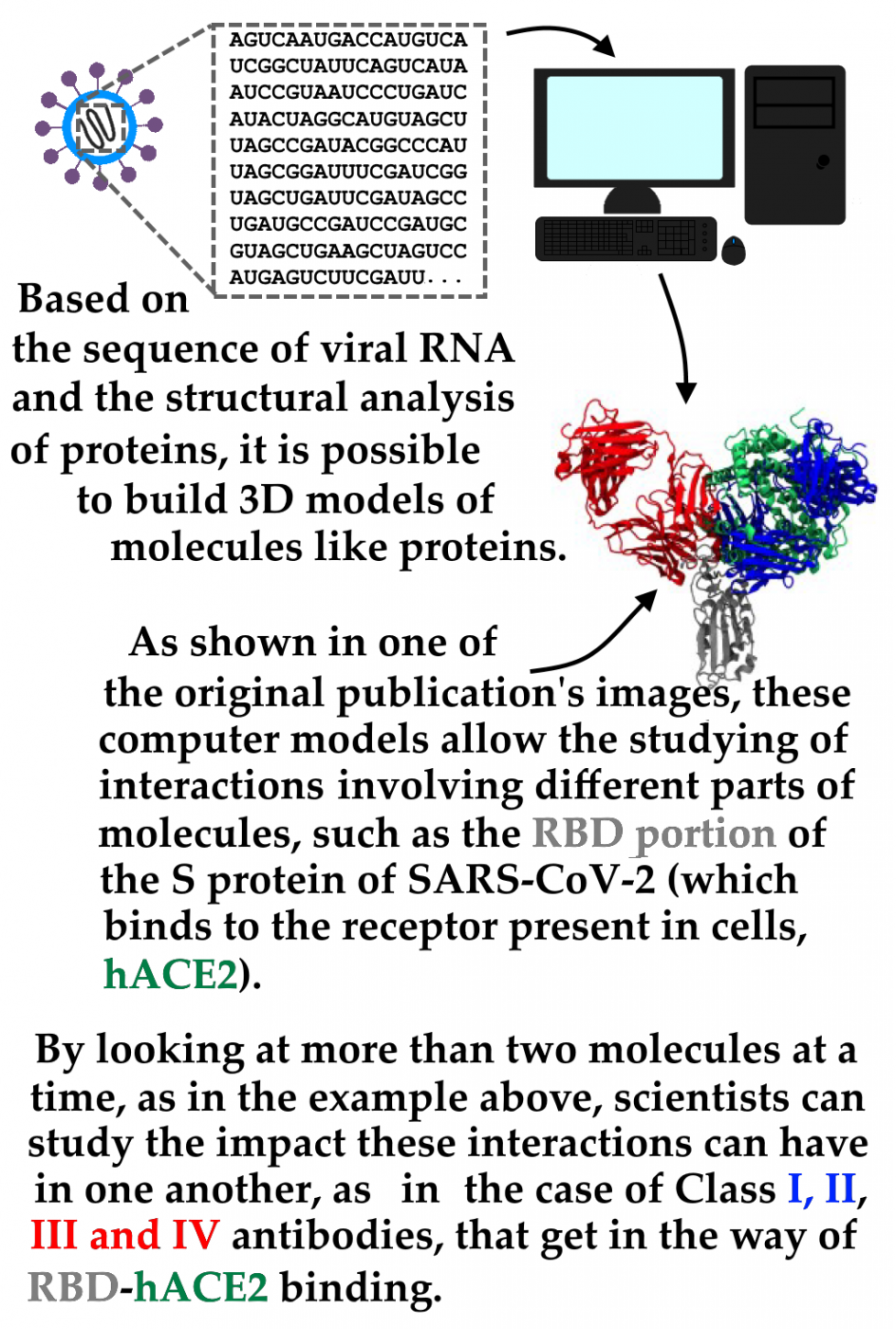
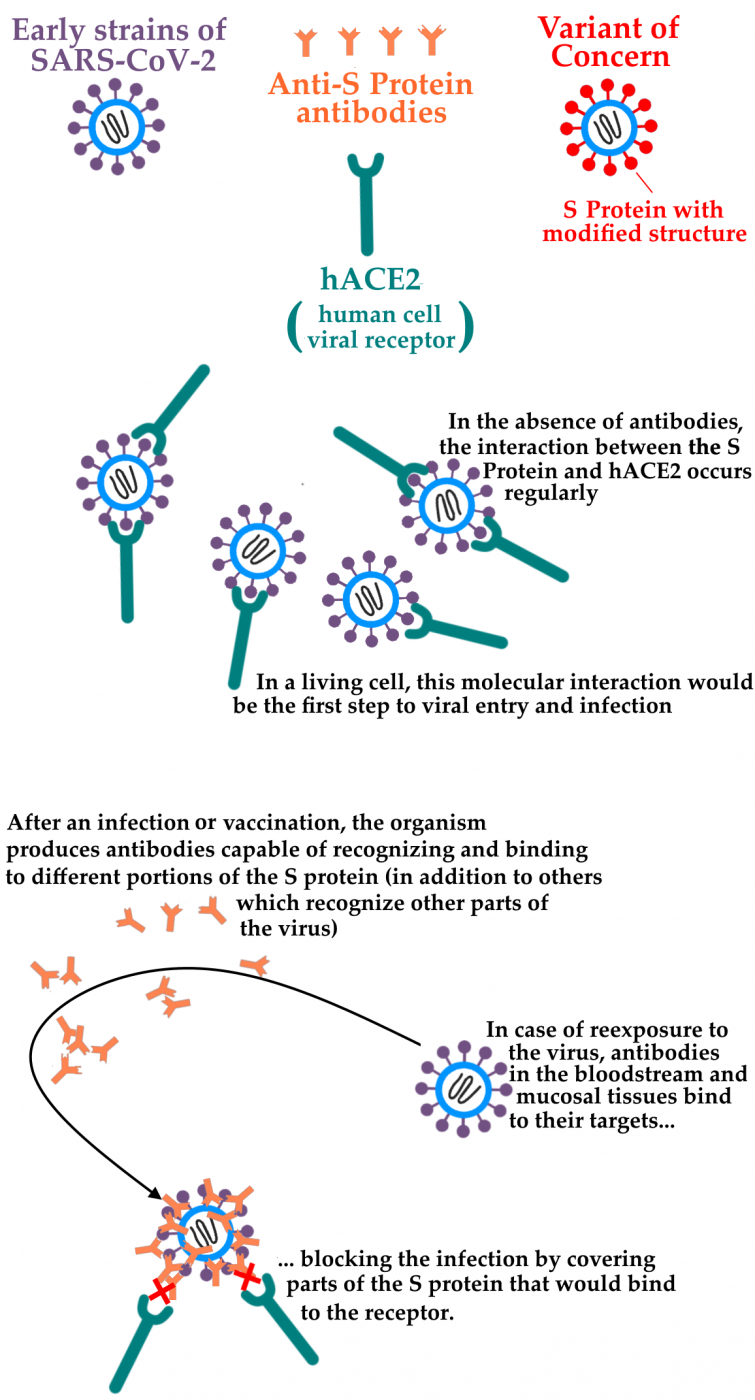
Of special interest to the research aimed at monitoring new variants is the study of the mutation in the viral genome (in particular, in the Spike glycoprotein gene, S protein), which promotes entry into human cells through interaction with Angiotensin Converting Enzyme 2 (hACE2), a molecule that acts as a receptor for the virus. The ability to bind to hACE2 may become greater or less according to changes in the S protein structure, which vary between Sars-CoV-2 strains and each of its variants. These changes in the protein S structure may also result in a decreased ability of antibodies generated in response to an infection - or possibly even after vaccination - to bind to protein S and neutralize the virus' ability to cause infection.
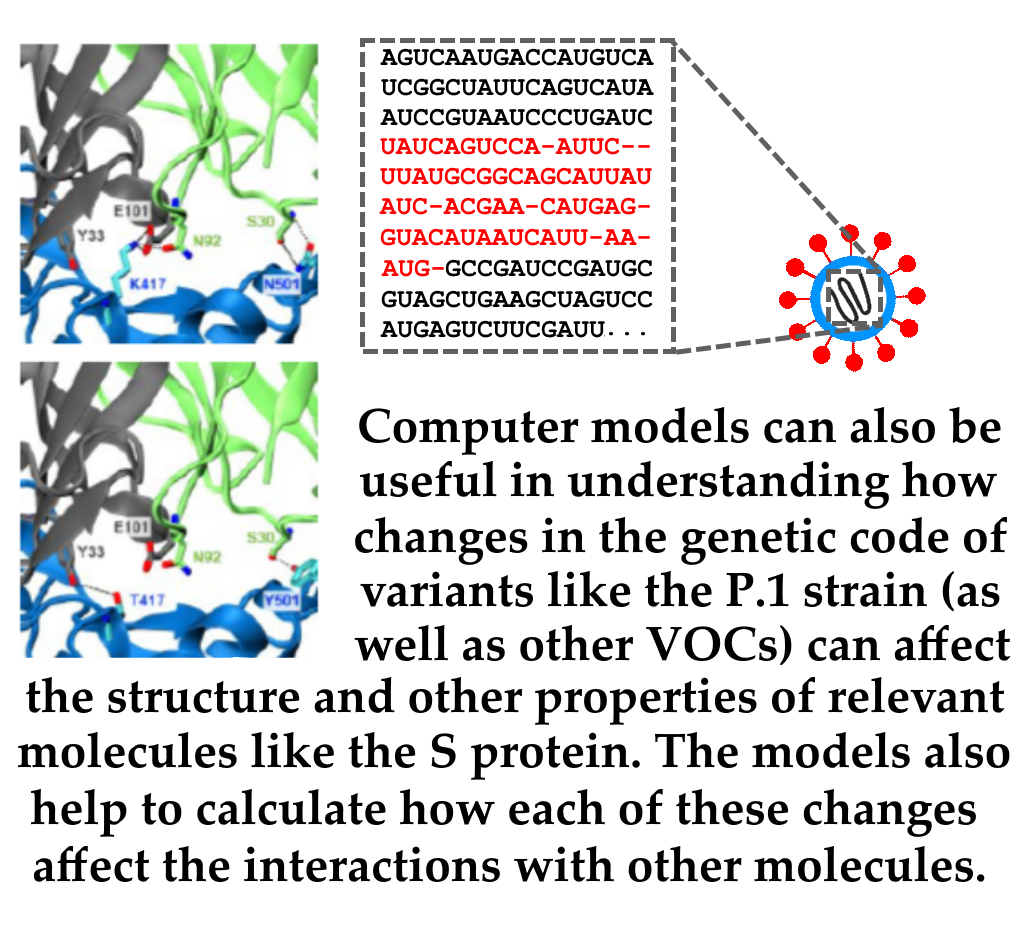
The present publication investigates these two possible S-protein mutations scenarios detected in variants such as P.1, B.1.351 strain variants, and B.1.1.7: that of a "stronger" interaction with the hACE2 receptor or that of a "weaker" interaction with anti-S-protein antibodies. The publication is the complete editorial process result, with review by independent researchers, of the preprint initially published by network researchers in March 2021. The paper describes a series of experiments performed by computational modeling of the molecules involved (hACE2, antibodies, and the protein S various "versions", referring to each of the variants studied).
Based on the computational simulation of the interaction between the molecules, it was possible to verify that altering the protein S structure had no marked effects on the interaction with the hACE2 receptor. On the other hand, when simulating the antibodies interaction generated in response to early Sars-CoV-2 strains with the S protein of the new variants, it was possible to see that there is a decrease in binding between the molecules, a finding that points to a potential for the immune response "escape". According to this hypothesis, the new variants would be more effective in evading the neutralization provided by antibodies, and this would be a more relevant mechanism to explain their rapid spread in the population.
Importantly, based on some measurements of affinity between variations of the S protein and the hACE2 receptor, other research groups have previously suggested that increased transmissibility would be related to a higher affinity between the viral protein and the human receptor, in contrast to the present study results. However, these studies have not explored the different S proteins interaction of concern variants with the neutralizing antibodies. Cases of variant reinfection have also recently been observed in patients who have recovered from previous infections, even though they are still producing antibodies. "This indicates that the escape from neutralization by antibodies is already a reality that influences the pandemic course", explains Lins. The paper also shows that the method used by the researchers is able to predict whether a new variant has the potential to be a variant of concern (VOC).